In Molecular epigenetics, we study the methylation of DNA and histone proteins as well as protein domains that specifically read these modifications. We investigate the mechanism and specificity of enzymes involved in epigenetic processes and are engaged in the development of enzyme inhibitors. In Synthetic biology, we aim to improve the properties of enzymes and proteins for various applications by rational and evolutionary design and develop artificial gene regulatory elements.
Member of the Stuttgart Research Center Systems Biology
Current research projects of Prof. Jeltsch
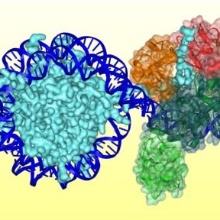
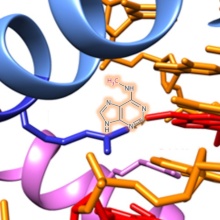
DNA methylation is an essential epigenetic modification involved in various biological processes. DNA methylation patterns are established in a largely unknown manner during the embryogenesis and development of mammals by the de novo DNA methyltransferases Dnmt3a and Dnmt3b. Dnmt1 methylates specifically hemimethylated CpG sequences after DNA replication, thereby copying DNA methylation patterns over cell divisions. We study the mechanism, regulation, and conformational transitions of human DNA methyltransferases in vitro and in cells. The aim of this project is to understand how DNA methyltransferases can fulfill their central cellular functions. Moreover, we devolp artifical, DNA methylaiton based epigenetic gene regulatory networks in bacteria for the memory and processing of external signals.
DNA methylation is an essential epigenetic modification involved in various biological processes. DNA methylation patterns are established in a largely unknown manner during the embryogenesis and development of mammals by the de novo DNA methyltransferases Dnmt3a and Dnmt3b. Dnmt1 methylates specifically hemimethylated CpG sequences after DNA replication, thereby copying DNA methylation patterns over cell divisions. We study the mechanism, regulation, and conformational transitions of human DNA methyltransferases in vitro and in cells. The aim of this project is to understand how DNA methyltransferases can fulfill their central cellular functions. Moreover, we devolp artifical, DNA methylaiton based epigenetic gene regulatory networks in bacteria for the memory and processing of external signals.
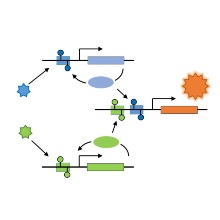
Recently, our group has described the design of synthetic-epigenetic switches that can capture transient environmental signals and store this information in cells in the form of DNA methylation patterns (Maier et al., Nat. Commun., 2017, 8: 15336). Potential applications of such synthetic epigenetic systems are diverse and involve the development of bacteria that could be used as living sensors to monitor cold chains, detect environmental pollution at production sites or nuclear power plants, or migrate through the human body to detect disease markers or metabolic states. In a new cooperation project supported by DFG, in cooperation with Prof. Radde (Institute of Systems Theory and Control) we want to improve the potential applicability of artificial epigenetic circuits and better understand their switching properties. Methods of synthetic biochemistry and quantitative dynamic modeling will be used to gain a mechanistic understanding of regulatory principles, information processing and epigenetic storage, increase their performance and complexity, and capture additional input signals.
Development and application of epigenome editing using locus specific chromatin modification enzymes
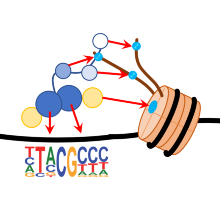
In this project, we are developing systems for epigenome editing that allow epigenetic modifications to be targeted at defined genomic loci. We employ chimeric enzymes in which artificial DNA recognition domains, e.g. Zinc finger proteins, TALs, or CRIPSR / Cas9 complexes for locus-specific DNA binding are fused to chromatin-modifying enzymes (such as DNA methyltransferases or lysine methyltransferases). We use Epigenom editing, in order to silence oncogenes in tumor cells or to correct epigenetic modifications in cells with imprinting defect. In addition, we study the changes of histone and DNA modifications to natural chromatin after the introduction of one or more defined modifications to better understand the process of reprogramming.
Post-translational modifications (PTMs) of N-terminal histone peptides play an important role in gene regulation and in the regulation of the state of chromatin. The biological effect of these PTMs is essentially mediated by protein interaction domains which bind PTM specifically to target proteins. The aim of this project is to identify and investigate binding domains for lysine methylation in histone and non-histone proteins. We plan to systematically clone and purify candidate domains and to investigate their interaction with modified histone peptides and with randomized peptides. We also develop binding domains for biotechnological applications and we apply reading domains to detect DNA methylation and histone modifications at genomic target loci in live cells.
In this project we investigate the enzymatic properties of Dnmts and PKMTs which contain cancer-mutations. Our results will help to define the carcinogenic effect of Dnmt and PKMT mutations observed in tumors and to understand how changes in DNA and protein methylation lead to cancer. This will support the development of more targeted therapies for tumors containing mutated Dnmts and PKMTs.
Current projects of Dr. Philipp Rathert
A central question of epigenome reserch is how chromatin or DNA binding proteins interact with one another to switch genes on or off at the right time point to establish and maintain a healthy cellular state. The project investigates the dynamics and wiring of chromatin regulators, as well as the induced transcriptional output under different conditions using state-of-the-art functional genetic tools (RNAi or CRISPR/Cas9) with cell biology and biochemical assays.
Further information on current projects of the group Rathert can be found here.
Research News
Kontakt

Albert Jeltsch
Prof. Dr.Head of Biochemistry Department and Acting Director of the IBTB