New publication in "Communications Biology"
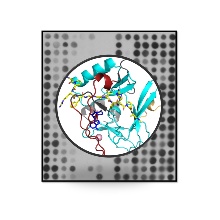
Methylation of lysine residues in proteins is a common post-translational protein modification with important biological functions. As a starting point for this work, we systematically analyzed the specificity of SETD2. SETD2 is a human protein lysine methyltransferase with potent disease compounds that methylates lysine 36 into histone H3 (H3K36). Using a peptide array based approach, we determined the specificity profile of SETD2 and identified new non-histone substrates of this enzyme. Using a peptide array based approach, we have determined the specificity profile of SETD2 and identified novel non-histone substrates of this enzyme. Strikingly, we observed that SETD2 prefers other amino acids than those present in the natural H3K36 sequence at several places and based on this we were able to design a SETD2 super-substrate that was methylated about 290-fold more strongly than H3K36, the best currently known SETD2 substrate. The very strong methylation of the super-substrate was observed on peptide arrays, with purified peptides, proteins in vitro and also in cells. We solved the structure of SETD2 bound to the super-substrate allowing to identify improved molecular interactions as potential reason for the increased methylation of the super-substrate, in addition to conformational changes, which provides novel insights into the mechanism of SETD2. Our data illustrate that substrate sequence design can strongly increase the activity of protein lysine methyltransferases.