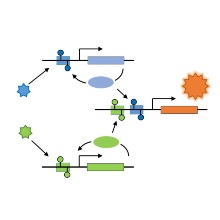
Recently, the group of Prof. Jeltsch has described the design of synthetic-epigenetic switches that can capture transient environmental signals and store this information in cells in the form of DNA methylation patterns (Maier et al., Nat. Commun., 2017, 8: 15336). Potential applications of such synthetic epigenetic systems are diverse and involve the development of bacteria that could be used as living sensors to monitor cold chains, detect environmental pollution at production sites or nuclear power plants, or migrate through the human body to detect disease markers or metabolic states. In a new cooperation project supported by DFG, the working groups of Prof. Jeltsch (Institute of Biochemistry and Technical Biochemistry) and Prof. Radde (Institute of Systems Theory and Control) want to improve the potential applicability of artificial epigenetic circuits and better understand their switching properties. Methods of synthetic biochemistry and quantitative dynamic modeling will be used to gain a mechanistic understanding of regulatory principles, information processing and epigenetic storage, increase their performance and complexity, and capture additional input signals.
Institute of Biochemistry and Technical Biochemistry, Department Biochemistry
Contact

Albert Jeltsch
Prof. Dr.Head of Biochemistry department and acting director IBTB